About Secmarker web server
The Secmarker web server was created by Didac Santesmasses, with support of Marco Mariotti. It provides online access to Secmarker, a selenocysteine tRNA (tRNA-Sec) identification tool. Secmarker was developed at Roderic Guigó lab at the CRG.
Citation:
Didac Santesmasses, Marco Mariotti and Roderic Guigó. Computational identification of the selenocysteine tRNA (tRNASec) in genomes. PLoS Comput Biol. 2017 Feb 13;13(2):e1005383. doi: 10.1371/journal.pcbi.1005383.Selenocysteine tRNA (tRNA-Sec)
Selenocysteine (Sec) is the 21st amino acid, a cysteine analogue with selenium replacing sulphur. Sec is inserted co-translationally in a small fraction of proteins called selenoproteins. Selenoproteins are present in the three domain of life: Eukarya, Bacteria and Archaea. Selenocysteine cognate tRNA (tRNA-Sec) is central to the selenoprotein synthesis process, playing a key role in both Sec biosynthesis and insertion. Sec is formed in a multistep process in which tRNA-Sec serves as a scaffold, so that Sec is synthesized already loaded on the tRNA. In selenoprotein synthesis, tRNA-Sec drives the re-coding of highly specific UGA codons from stop signals to Sec. Not all organisms use Sec. In selenoprotein-devoid organisms, tRNA-Sec is absent. Identification of tRNA-Sec in a genome can be used as marker for the Sec utilization trait.
The anticodon in tRNA-Sec is the UCA, (complementary to UGA). Like all tRNAs, its secondary structure comprises (Fig 1) an aminoacyl acceptor arm (A-stem), a dihydrouridine arm (D-stem and D-loop), an anticodon arm (C-stem and C-loop), a variable arm (V-stem and V-loop) and a TψC arm (T-stem and T-loop). It is the longest tRNA, with 90-100 nucleotides, rather than the conventional ~75 nucleotides in canonical tRNAs [1]. It has an unusual structure, different from the canonical 7/5 in other tRNAs (where 7 and 5 are the number of base pairs (bp) in the A and T stems, respectively). The tRNA-Sec adopts a 9/4 fold in eukaryotes and archaea, and a 8/5 fold in bacteria [2]. The acceptor and T arms have 13 bp in total, compared to 12 bp in the usual 7/5 structure in other tRNAs. It has an exceptionally long variable arm, even longer than those of type-2 tRNAs (e.g. tRNA-Ser) [3]. The D arm of tRNA-Sec has a long D-stem, with 6 bp in eukaryotes and bacteria, and 7 bp in archaea [4], and a 4 bp D-loop, in contrast to the 3-4 bp D-stem and 7-12 nt D-loop in the canonical tRNAs [3]. The unique structure of tRNA-Sec allows it to be recognized by SerRS, like tRNA-Ser, while conversely acting as the exclusive target of PSTK, SecS (or SelA), and EF-Sec (or SelB): SerRS recognizes the variable arm of tRNA-Sec, similar to the long variable arm in tRNA-Ser, while the Sec synthesis factors must strictly discriminate the tRNA-Sec. In contrast, PSTK phosphorylates tRNA-Sec but not tRNA-Ser by recognizing the tRNA-Sec D arm [5], and SecS contacts the characteristic 13 bp AT-stem [6].
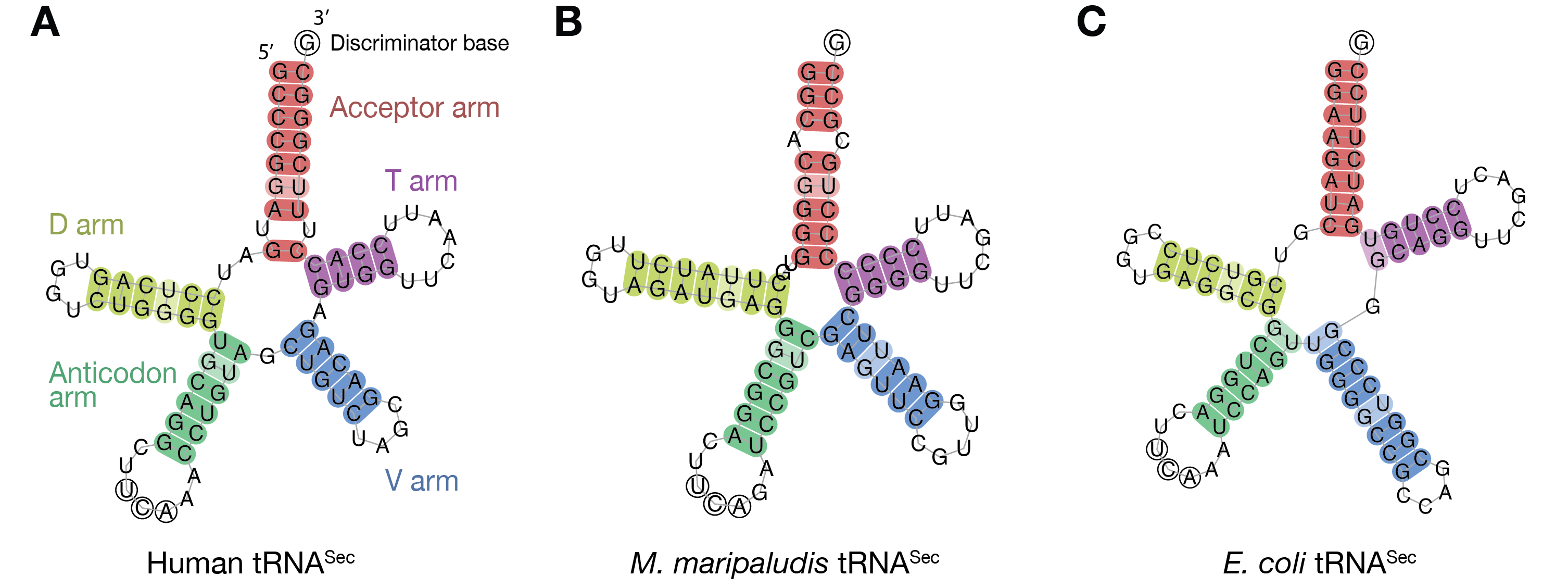
Fig 1. Structure-based cloverleaf models of tRNA-Sec. (A) Homo sapiens (Eukarya). (B) Methanococcus maripaludis (Archaea). (C) Escherichia coli (Bacteria). The acceptor arm, D arm, anticodon arm, variable arm and T arm are colored red, yellow, green, blue and purple, respectively. The anticodon UCA is indicated with circled residues.
References
2. Hubert N, Sturchler C, Westhof E, Carbon P, Krol A. The 9/4 secondary structure of eukaryotic selenocysteine tRNA: more pieces of evidence. RNA (New York, NY). 1998 Sep;4(9):1029-33.
3. Itoh Y, Sekine Si, Suetsugu S, Yokoyama S. Tertiary structure of bacterial selenocysteine tRNA. Nucleic acids research. 2013 Jul;41(13):6729-38.
4. Sherrer RL, Araiso Y, Aldag C, Ishitani R, Ho JML, S ̈oll D, et al. C-terminal domain of archaeal O-phosphoseryl-tRNA kinase displays large-scale motion to bind the 7-bp D-stem of archaeal tRNA(Sec). Nucleic acids research. 2011 Feb;39(3):1034-41.
5. Chiba S, Itoh Y, Sekine Si, Yokoyama S. Structural basis for the major role of O-phosphoseryl-tRNA kinase in the UGA-specific encoding of selenocysteine. Molecular cell. 2010 Aug;39(3):410-20.
6. Palioura S, Sherrer RL, Steitz TA, Söll D, Simonovic M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science (New York, NY). 2009 Jul;325(5938):321-5.